Translating a BEP Document into Schema Code¶
One of the most important steps of a BEP is translating it from a plain text (google/microsoft document) as a yaml schema. The yaml schema necessary for the following steps:
render the spec into its ultimate compiled form
apply and validate the new rules and requirements introduced by the BEP
merge their BEP into the BIDS Spec
For the purpose of this guide we’ll be referencing a completed BEP starting from its initial Google doc form and finishing with its translation into schema.
The Google Doc¶

Defining new file names¶
BEP 009 introduced files of the form
sub-<label>/[ses-<label>/]pet/sub-<label>[_ses-<label>][_task-<label>][_trc-<label>][_rec-<label>][_run-<index>]_pet.nii[.gz]
This path consists of the following objects, which are defined in the schema:
Object type |
Description |
Example |
Defined in |
Introduced by BEP009 |
|---|---|---|---|---|
entity |
key-value pairs |
|
|
|
data type |
directory describing data |
|
|
|
suffix |
end of the file stem (before extension) |
|
|
|
extension |
file type indicator |
|
none |
BIDS also has the concept of a “modality”, which does not always appear in the path, but is determined by the data type.
Object type |
Description |
Example |
Defined in |
Introduced by BEP009 |
|---|---|---|---|---|
modality |
primary recording technique |
|
|
Datatypes and modalities¶
PET introduces a new datatype (pet/) and modality (pet).
For new modalities the first step should be to add that modality to modalities.yaml.
# src/schema/objects/modalities.yaml
---
# This file describes modalities supported by BIDS.
mri:
display_name: Magnetic Resonance Imaging
description: |
Data acquired with an MRI scanner.
[...]
+pet:
+ display_name: Positron Emission Tomography
+ description: |
+ Data acquired with PET.
[...]
mrs:
display_name: Magnetic Resonance Spectroscopy
description: Data acquired with MRS.
Likewise, update datatypes.yaml:
# src/schema/objects/datatypes.yaml
---
# This file defines valid BIDS datatypes.
anat:
value: anat
display_name: Anatomical Magnetic Resonance Imaging
description: |
Magnetic resonance imaging sequences designed to characterize static, anatomical features.
[...]
+pet:
+ value: pet
+ display_name: Positron Emission Tomography
+ description: |
+ Positron emission tomography data
[...]
nirs:
value: nirs
display_name: Near-Infrared Spectroscopy
description: Near-Infrared Spectroscopy data organized around the SNIRF format
Here, the value field indicates the string that will appear in the file name.
Finally, rules/modalities.yaml must be updated to associate the new
datatype to the new modality:
# src/schema/rules/modalities.yaml
---
mri:
datatypes:
- anat
- dwi
- fmap
- func
[...]
+pet:
+ datatypes:
+ - pet
[...]
mrs:
datatypes:
- mrs
Suffixes¶
To add a suffix, update suffixes.yaml:
---
# This file defines valid BIDS file suffixes.
# For rules regarding how suffixes relate to datatypes, see files in `rules/files/raw/`.
TwoPE:
value: 2PE
display_name: 2-photon excitation microscopy
description: |
2-photon excitation microscopy imaging data
[...]
+pet:
+ value: pet
+ display_name: Positron Emission Tomography
+ description: |
+ PET imaging data SHOULD be stored in 4D
+ (or 3D, if only one volume was acquired) NIfTI files with the `_pet` suffix.
+ Volumes MUST be stored in chronological order
+ (the order they were acquired in).
[...]
unloc:
value: unloc
display_name: Unlocalized spectroscopy
description: |
MRS acquisitions run without localization.
This includes signals detected using coil sensitivity only.
Entities¶
To add an entity, entities.yaml must be updated.
In this case, PET introduced a trc-<label> entity, short for “tracer”:
---
# This file describes entities present in BIDS filenames.
acquisition:
name: acq
display_name: Acquisition
description: ...
type: string
format: label
[...]
+tracer:
+ name: trc
+ display_name: Tracer
+ description: |
+ The `trc-<label>` entity can be used to distinguish sequences using different tracers.
+
+ This entity represents the `"TracerName"` metadata field.
+ Therefore, if the `trc-<label>` entity is present in a filename,
+ `"TracerName"` MUST be defined in the associated metadata.
+ Please note that the `<label>` does not need to match the actual value of the field.
+ type: string
+ format: label
[...]
tracksys:
name: tracksys
display_name: Tracking System
description: ...
type: string
format: label
Note that the name is what shows up in filenames before a dash (-).
This is often a shortened version of the reference name, in this case, tracer.
The display_name is a name that might be used when creating a document
or entry form for presenting or requesting the entity value.
type and format indicate the range of acceptable values.
For entities, type is always string,
and format may be label (alphanumeric) or index (numeric).
Extensions¶
It is rare, but adding a new extension entails updating extensions.yaml.
nii:
value: .nii
display_name: NIfTI
description: |
A Neuroimaging Informatics Technology Initiative (NIfTI) data file.
nii_gz:
value: .nii.gz
display_name: Compressed NIfTI
description: |
A compressed Neuroimaging Informatics Technology Initiative (NIfTI) data file.
As with datatype and suffix, the string that appears in the file path is in the value field.
File naming rules¶
Your next steps are to move from this template file name as described in your document to formally defining those entities within the schema.
The definitions for these file entities are located within src/schema/rules/files.
Since this BEP is referring to raw data we go one level further to src/schema/rules/files/raw and create a new pet.yaml file to record each entity/rule.
There are additional directories for common elements (common) and derivative files (derivatives/), but we’re only focusing on raw PET data for this example.
$ tree src/schema/rules/files -L 1
src/schema/rules/files
├── common
├── deriv
└── raw
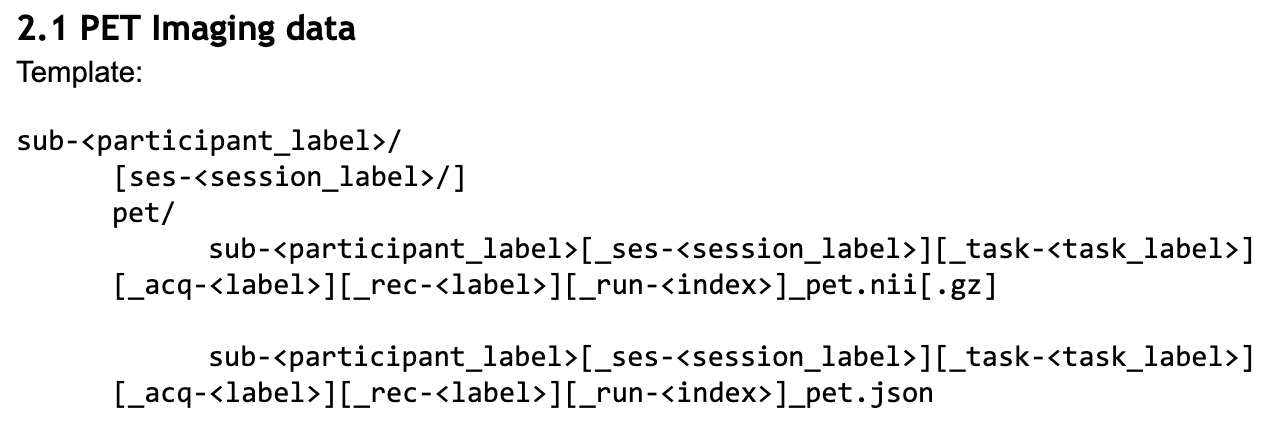
Given the template above we want to mark all entities surrounded by [] as optional while everything else not contained in [] will be marked as required.
(The acquisition entity was dropped in favor of tracer after the Google Doc was closed to edits, before the final inclusion of BEP 009.)
So, we create that pet.yaml file and begin to populate it.
# src/schema/rules/files/pet.yaml
---
pet:
suffixes:
- pet
extensions:
- .nii.gz
- .nii
- .json
datatypes:
- pet
entities:
subject: required
session: optional
task: optional
tracer: optional
reconstruction: optional
run: optional
In rare cases, new files may not have any entities. See Filename construction rules for more details.
Sidecars and Metadata Rules¶